Investigating the role of new regulators of mRNA translation: case study in the hetero- and autotroph model bacteria, E. coli and Synechocystis
PI: Grégory Boel
Postdoc length: 2 years
Work place: Microbial Gene Expression Laboratory
Contact: Grégory Boël (boel[at]ibpc.fr)
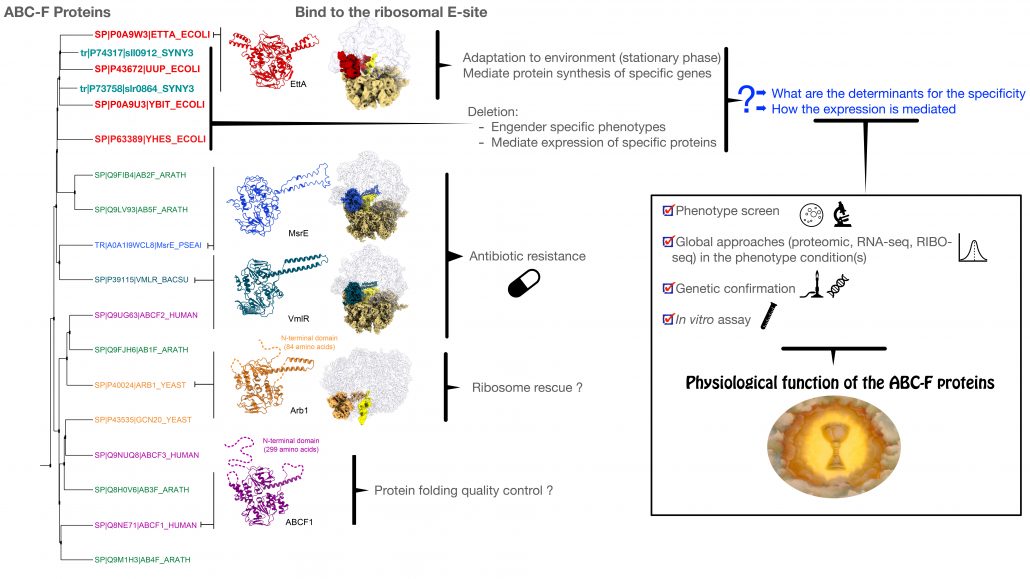
Protein synthesis is at the heart of the central dogma of biology. Even if this mechanism can be reconstructed in vitro with a minimal set of translation factors and ribosomes, the efficiency is far lower than what happens in vivo, demonstrating that our understanding of the translation apparatus is incomplete. A few years ago we identified that members of the ABC-F protein family work as translation factors and that they are present ubiquitously in prokaryote and eukaryote (1-3). Multiple occurrences of ABC-F are often found in a genome suggesting different functions for each paralog (3).
Some of these ABC-F proteins are involved in antibiotic resistance (4) but the majority play a role in the regulation of mRNA translation. We have identified and begun the characterization of all of the ABC-F proteins in 2 model bacteria: the proteobacterium Escherichia coli and the cyanobacterium Synechocystis. We have shown that, in both organisms, ABC-F are important for the adaptation of the organism to its environment (unpublished), but the specificity of each paralog remains to be characterized in detail.
The successful candidate will conduct a research program around the functional and physiological characterization some of the ABC-F proteins in cyanobacteria and E. coli. He/She will benefit from the multiscale investigations (proteomic, RNAseq and structural characterization) that the lab has done during the last 5 years. The study will also take advantage of the methods that the lab has implemented in terms of protein expression (5, 6) and from the specific expertise in cryo-EM or computational biology and modelling from our collaborators in France and in the US.
We are looking for a highly motivated candidate with a main expertise in all the fields of microbiology and molecular biology. An expertise in high throughput sequencing (mRNA-seq and / or ribo- seq) would be a plus. The candidate should be able to carry out a research program with some independence and have good communication skills. Attentive to the academic career development of its members, the lab will strongly support the candidate to present the CNRS national competition for permanent position.
1- Fostier CR, Monlezun L, Ousalem F, Singh S, Hunt JF, Boël G. ABC-F translation factors: from antibiotic resistance to immune response. FEBS Lett. 2021 Mar;595(6):675-706. doi: 10.1002/1873-3468.13984
2- Boël G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, Testa AJ, Fischer JJ, Wieden HJ, Frank J, Gonzalez RL Jr, Hunt JF. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol. 2014 Feb;21(2):143-51. doi: 10.1038/nsmb.2740.
3- Chen B, Boël G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL Jr, Hunt JF, Frank J. EttA regulates translation by binding the ribosomal E site and restricting ribosome-tRNA dynamics. Nat Struct Mol Biol. 2014 Feb;21(2):152-9. doi: 10.1038/nsmb.2741
4 – Fostier C. R., Ousalem F., Leroy E. C., Ngo S., Soufari H, Innis A, Hashem Y., Boël G. Regulation and mechanistic basis of macrolide resistance by the ABC-F ATPase MsrD. bioRxiv 2021.11.29.470318; doi: 10.1101/2021.11.29.470318
5 – Boël G, Letso R, Neely H, Price WN, Wong KH, Su M, Luff J, Valecha M, Everett JK, Acton TB, Xiao R, Montelione GT, Aalberts DP, Hunt JF. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016 Jan 21;529(7586):358-363. doi: 10.1038/nature16509
6 – Lipońska A, Ousalem F, Aalberts DP, Hunt JF, Boël G. The new strategies to overcome challenges in protein production in bacteria. Microb Biotechnol. 2019 Jan;12(1):44-47. doi: 10.1111/1751-7915.13338.