Research Themes
Task 1: The evolution of energy-producing organelles from bacterial ancestors
(PARTNERS 1, 2, 3, 4, 6, 7)
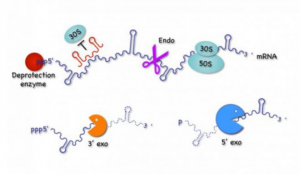
Modern chloroplasts (cp) and mitochondria (mt) have tiny genomes that only code for a fraction of the factors required for their own gene expression and function. Nevertheless, the organellar machineries for transcription, translation, and mRNA decay have a pronounced prokaryotic nature, because most of the factors involved are the products of ancestral bacterial genes that have been transferred to the nucleus. The nucleus in turn tightly controls the expression of organellar genes by encoding gene-specific factors that bind organellar mRNAs and govern their maturation/stability and translation.
The main goal of Task 1 is to improve our understanding of the evolutionary adaption that had to occur for the ancient bacterial ancestors of cp and mt to survive as endosymbionts within their new hosts. Our goal is to understand the very early events that led to the initial bacterial survival within the host, in particular the reappropriation of antimicrobial peptides to function as transit peptides used to bring proteins into modern day organelles. In parallel, we are studying the later evolutionary events leading to the institution of the post-transcriptional control mechanisms that allowed endosymbionts to transfer most of their genes to the nucleus of the host cell, through the study of the evolution of OPR/PPR proteins involved in regulated translation and mRNA stability in micro-algae. We are also studying new factors involved in translation and RNA maturation/decay in bacteria and cp, as well as new RNA-based mechanisms of control of gene expression, in particular those encoding membrane proteins. Lastly, we are studying the cost of maintaining organellar activity to the host cell in the case of yeast S. cerevisiae mt by examining the link between oxidative stress, post-transcriptional gene regulation and telomere length.
DYNAMO has been highly successful in terms of our overall accomplishments for Task 1, with 103 publications in total so far and the establishment of new collaborations between the partners that can be directly credited to the project.
Task 2: Membrane dynamics and their regulation across evolution
(PARTNERS 1, 2, 4, 5, 7)
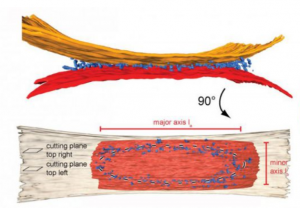
Membrane structure and dynamics are key to life. They afforded a rapid increase in evolution rate following the emergence of the first bio-active molecules from the primordial soup by allowing protection from the environment, partitioning of primitive cell functions such as nucleic acid replication, and the early development of mechanisms to generate and store energy.
Our goal in Task 2 is to characterize the MP-protein and MP-lipid interactions that ultimately shape membrane biogenesis, dynamics and the regulation of their morphologies in mt, in cp and in bacteria. In mt, we are particularly interested in dissecting the molecular and structural mechanism of mitochondrial fusion by Dynamin related proteins. In cp, we are focusing on how microalgae display a variety of previously unknown strategies to optimize photosynthesis and light perception in aquatic environments. In bacteria, our studies of the AtpF-dependent membrane proliferating model system extend in several directions, including AtpF orthologs from Synechocystis or Chlamydomonas cp. In parallel, we are studying the stabilization and structural biology of MPs and their complexes involved in membrane transport and signalling, exploiting our expertise in surfactant chemistry. We will expand on our ongoing success in DYNAMO task2 with 68 publications in total so far and numerous collaborations.
Task 3: Supramolecular organisation of electron transfer
(PARTNERS 1, 2, 4, 5, 6, 7)
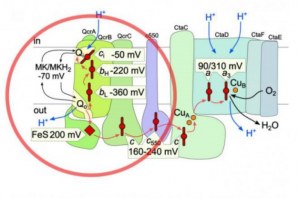
Life crucially depends on energy harvesting. Most organisms use a unique mechanism that relies on optimised electron flow to drive photosynthesis or respiration.
The aim of Task 3 is to study the organization and function of the electron transfer chains (ETC) in energy-transducing membranes, from the atomic level to the supramolecular organization of complexes. This includes the control of energy input, as cells need to harvest sufficient energy without overloading the ETC, which is particularly crucial in photosynthesis where light intensity is highly variable. We are studying the sophisticated mechanisms evolved by plants and algae, variable among clades, to cope with these fluctuations, optimize the photosynthetic electron flow or dissipate excess energy. We are also studying the electron bifurcation steps in ETC, such as the Qo cycle at the level of the Rieske/cytochrome b complex, that improves ETC efficiency by increasing the yield of translocated protons per transferred electron, while introducing a rate-limiting step susceptible to promote reactive oxygen species (ROS) formation. Quinones are key electron transporters of the ETC, acting upstream of the bifurcation step. We are characterizing the poorly known quinone biosynthetic pathway, paving the way for the development of therapeutic drugs for genetic quinone deficiencies or engineering bacteria to produce various quinone analogs for structure-function studies of respiratory complexes. We are developing and using innovative functional and structural approaches to address these issues and ultimately foster new ways to harvest energy, such as bio-inspired molecular catalysts for water oxidation and CO2 reduction, as well as bio-photovoltaic cells based on living microalgae. These major challenges that address the growing demand for both food and energy and help mitigate the consequences of climate change